Mophing
We devise computational algorithms to characterize folding pathways and strengthen the bridge between protein sequence and its structure.
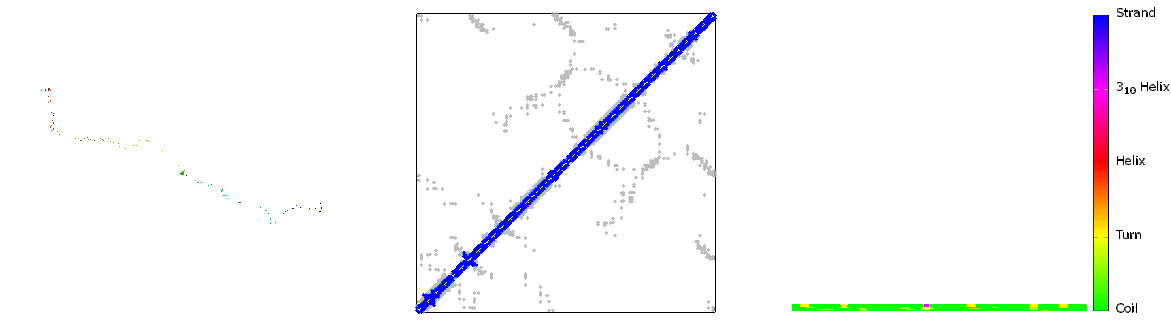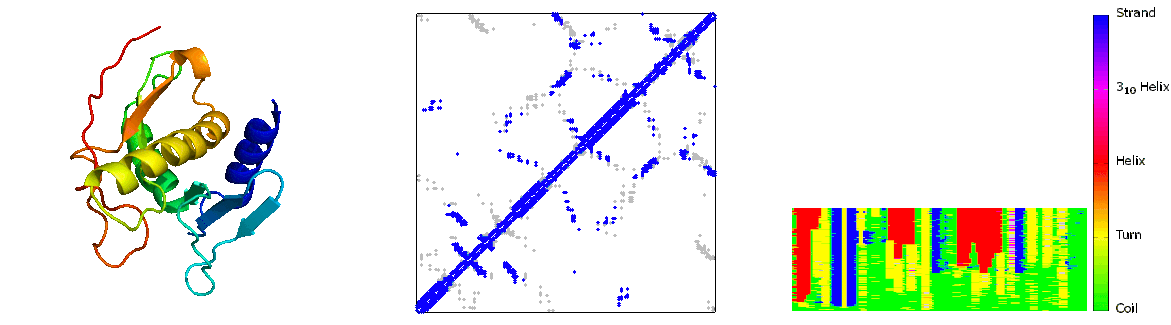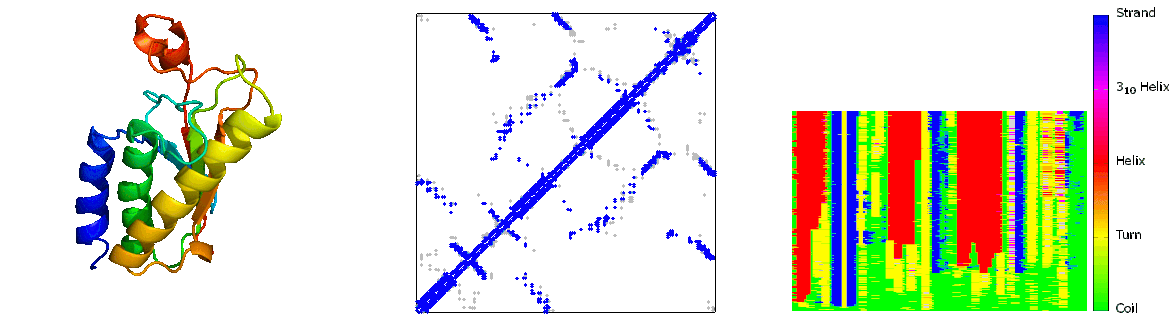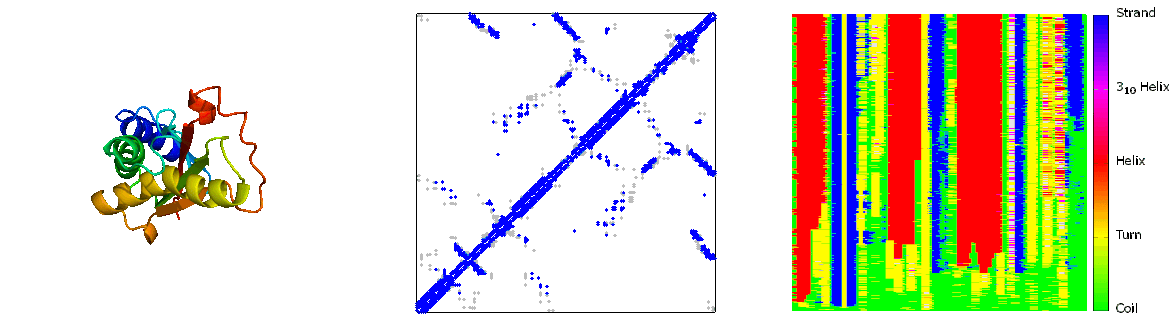
What are folding pathways?
Protein folding pathways outline the step-by-step process through which a sequence of amino acids folds into its functional three-dimensional structure, known as the native state. This process involves the sequential formation of secondary structures like alpha helices and beta sheets, driven by hydrogen bonding among backbone atoms. These structures then fold further into the protein's tertiary structure, stabilized by interactions such as hydrophobic forces, electrostatic interactions, and disulfide bonds. The folding pathway is influenced by the protein's primary sequence and environmental factors like temperature and pH. Molecular chaperones assist in correct folding by preventing misfolding and aggregation. Understanding protein folding pathways is crucial for elucidating protein function, structure-function relationships, and the development of therapies for diseases related to protein misfolding, such as Alzheimer's and Parkinson's diseases. Experimental techniques such as NMR spectroscopy and computational modeling are employed to study these pathways and unravel the complexities of protein folding mechanisms.