PhiGnet
Leveraging physics-informed graph neural networks, we seek to annotate protein functions with greater accuracy and interpretability, extending even to the residue level.
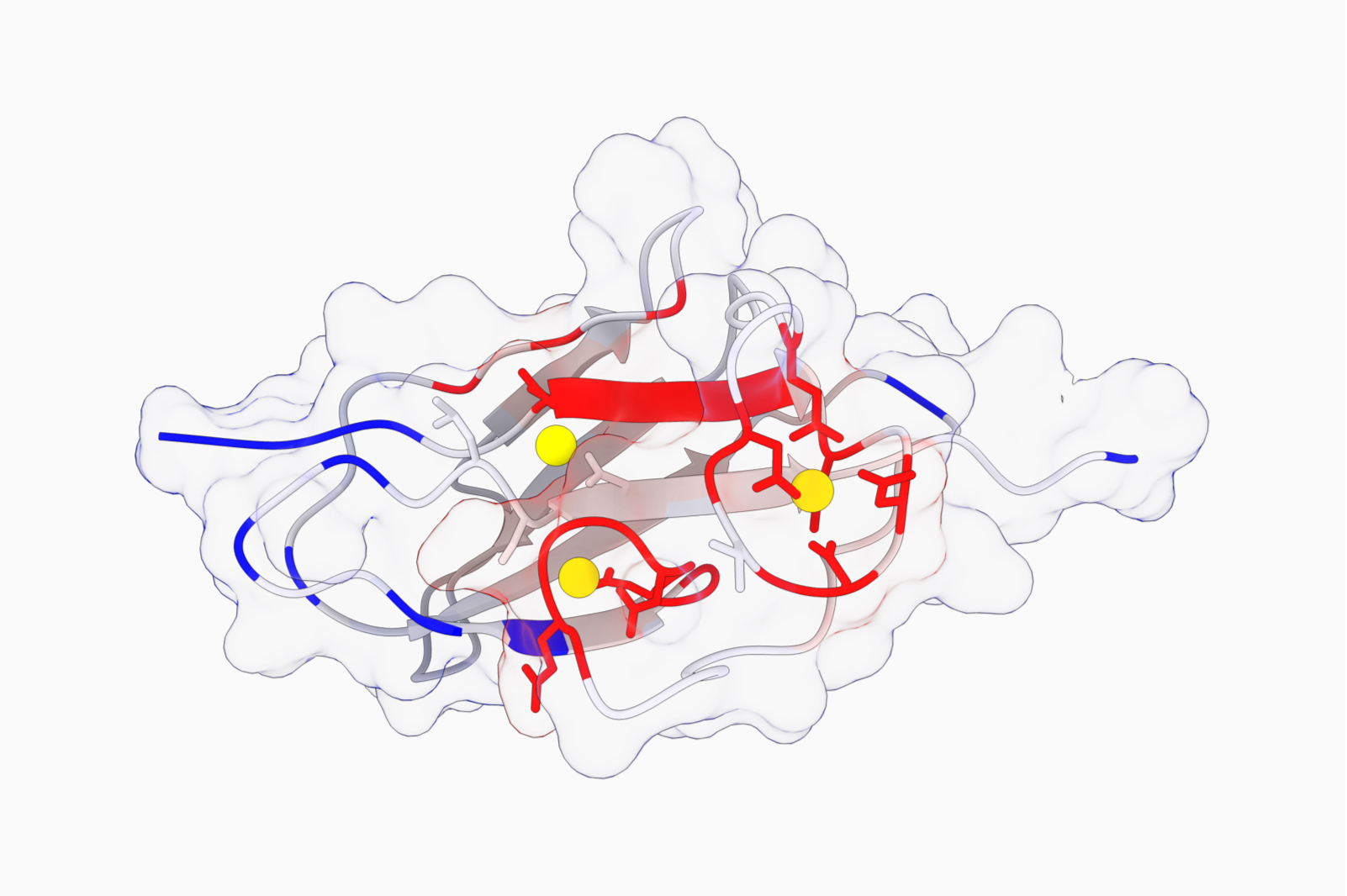
How to annotate protein function using PhiGnet?
An amino acid sequence encapsulates the evolutionary interactions between its constituent amino acids, dictating the rules governing the stabilization of protein folds and functions. While essential, delineating the precise interactions necessary and sufficient for structure and biochemical function poses a challenge due to the intricate nature of cooperative interactions. In this endeavor, we seek to statistically define these rules for the assignment of functional annotations to proteins, harnessing evolutionary data extracted from multiple sequence alignments in the absence of structural information.