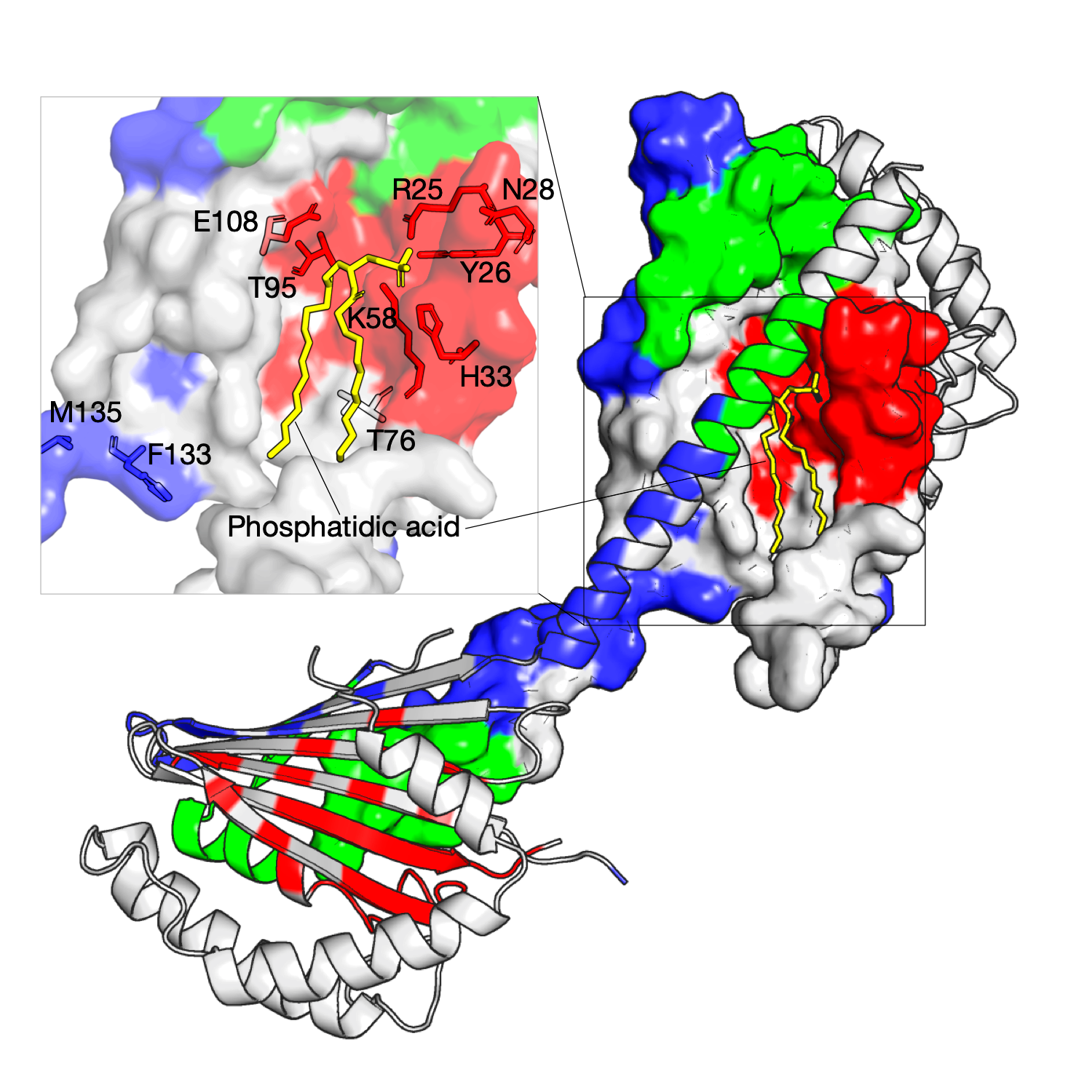
Structural determinants of lipid specificity
The Ups/PRELI lipid transfer proteins are widely conserved in eukaryotes and can be classified into two subfamilies by sequence similarity (Ups1/PRELID1 and Ups2/PRELID3. The Ups1–Mdm35 complex directly transfers phosphatidic acid (PA) between mitochondrial membranes allowing cardiolipin (CL) synthesis in the inner membrane. The gain-of-function Ups1 mutants from this approach can be categorised into two different functional regions: (a) residues facing to the inner substrate-binding cavity that tailor the cavity for selective accommodation of PA (K58, T95, E108) and (b) residues proximal to the Ω loop (K58, T76) and C-terminal α3 helix that likely have a role together in lipid extraction and capping an occupied binding cavity (F133, M135)[1]. As shown in the right figure, three residue communities are inferred from homologous sequences of the Ups1, and these residues in red are identified and mapped on the structure (PDB: 4YTX, chain A, B, M, N), as shown, they are located at the tunnel-like binding cavity within the hydrophobic core.
2020-10-13
Residue communities, Ups/PRELI, Ups1/PRELID1, Ups2/PRELID3
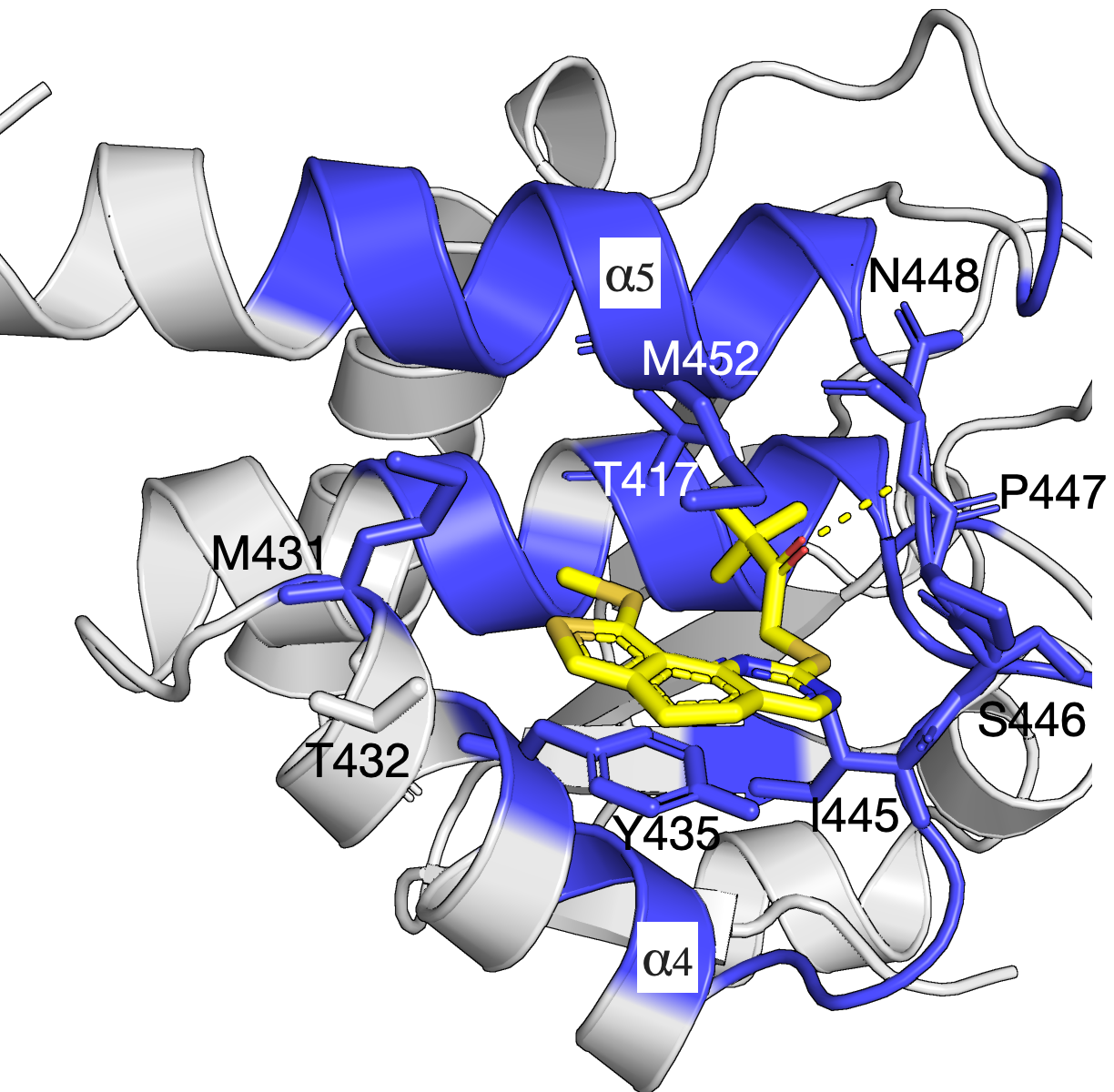
Network of residues in the allosteric pocket govern Compound 1 inhibition and MKP5 selectivity
The mitogen-activated protein kinases (MAPKs) are a family of serine/threonine kinases that play indispensable roles in signal transduction pathways that control a plethora of physiological responses[1]. As shown in the left figure, an experimentally demonstrated allosteric binding pocket[1] is identified by our SAEC method. In the analysis, only the homolog sequences are used to compute the evolutionary information and infer the binding pockect that agrees with that demonstrated in experiment. The identified network of residues is shown in blue and reveals key interactions between Compound 1 and MKP5-CD (PDB: 6MC1).
Antagonistic activity on floral regulators FT and TFL1
The His88 and Asp144 residues, which form a hydrogen bond in TFL1, but not FT, and which are likely the most critical residues for distinguishing FT and TFL1 activity[1].
2020-09-18
Residue communities, functional networks, TFL1, FT
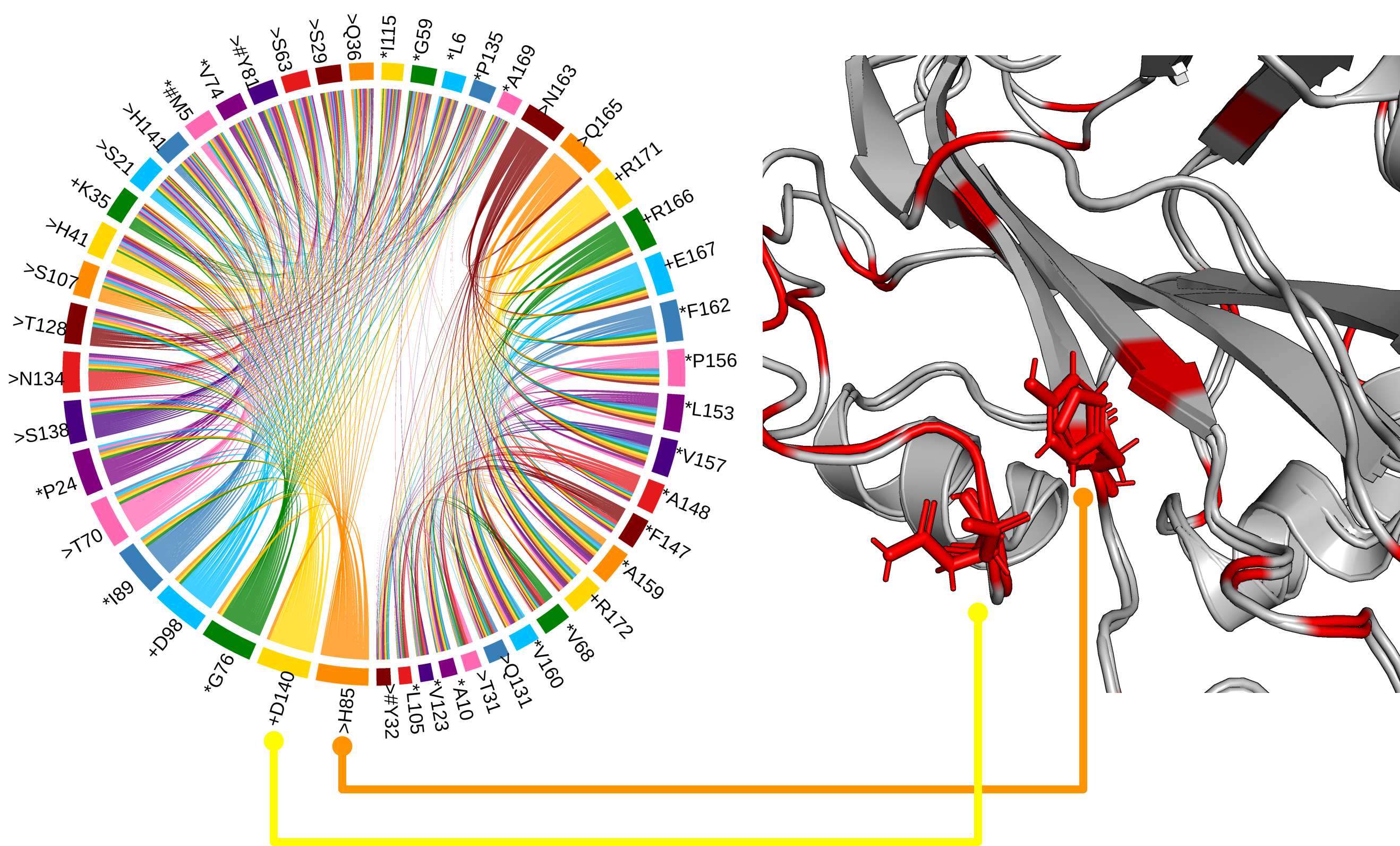
LetB tunnel is critical for function
LetB forms a periplasm-spanning tunnel consisting of seven stacked rings and provides a hydrophobic pathway for the translocation of lipids across the periplasm. Rings 1, 5, 6, and 7 are dynamic and adopt open and closed states. Our method has successfully identified functional communities in the LetB (PDB: 6V0C) as shown in the figure (left). The pore-facing residues in red are inferred using AmoAi and contribute to the formation of the tunnel that mediates lipid transport in different states.
2020-08-27
Residue communities, lipid transportation, LetB

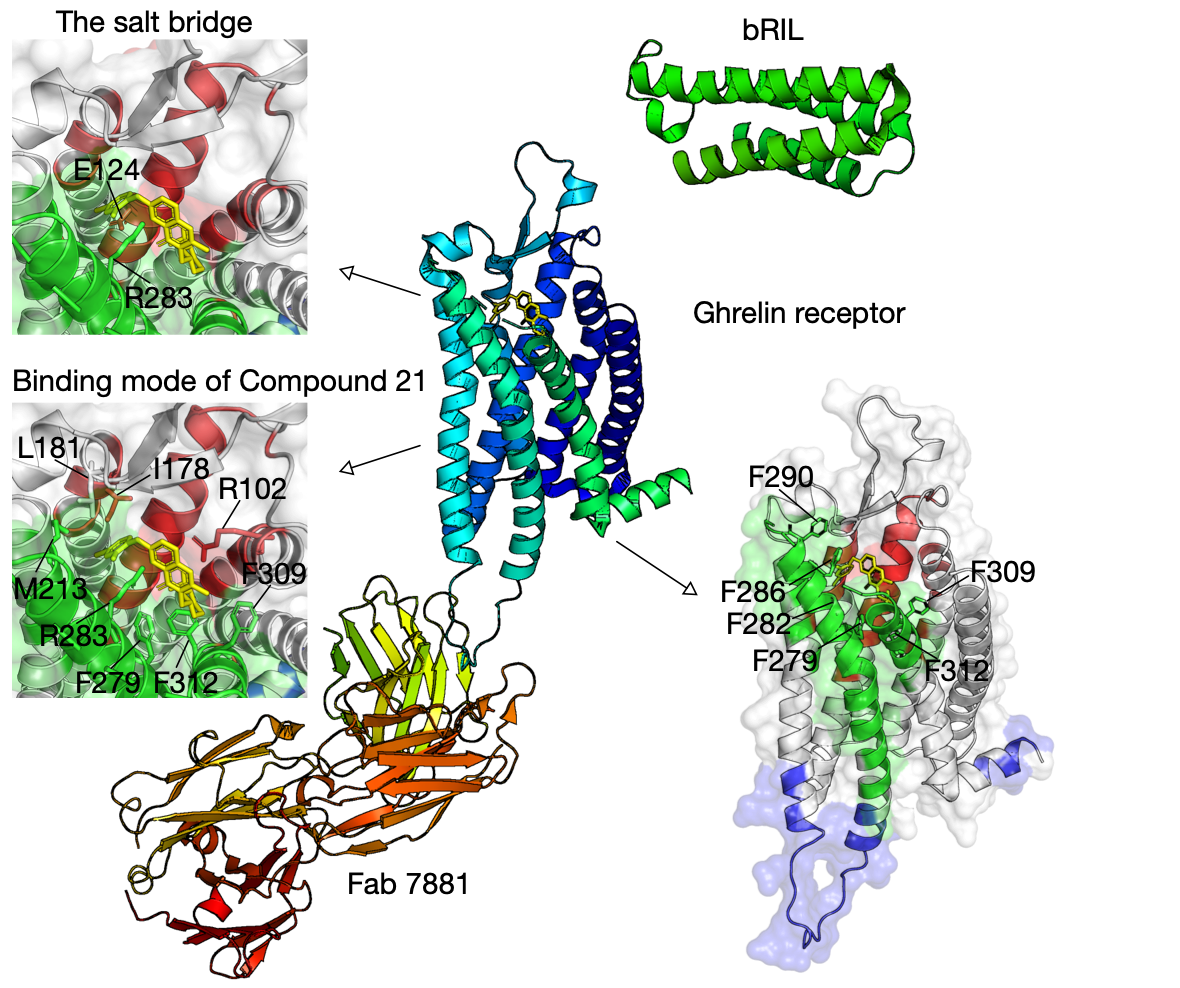
Bifurcated ligand-binding pocket in the ghrelin receptor
Ghrelin is a gastric peptide hormone with important physiological functions. The unique feature of ghrelin is its Serine 3 acyl-modification, which is essential for ghrelin’s activity. Our method has successfully identified functional communities that cover the ligand-binding pocket of the ghrelin receptor (hGHSR PDB: 6KO5), e.g., bifurcated by a salt bridge between E124 and R283.
2020-08-25
Residue communities, Ghrelin receptor, GPCR
The chorismate mutase
The chorismate mutase is an enzyme that is only found in fungi, bacteria, and higher plants, and it catalyzes the chemical reaction of converting chorismate to prephenate at the central branch-point of the shikimate pathway. As a clssical system, it has been used to understand principles of catalysis and enzyme design. Our method has successfully identified functional communities in the chorismate mutase (PDB: 1ECM) as shown in the figure (left). The residues that are functionally relevant to the catalytic activity are captured in the community (blue), and they are shown in stick.
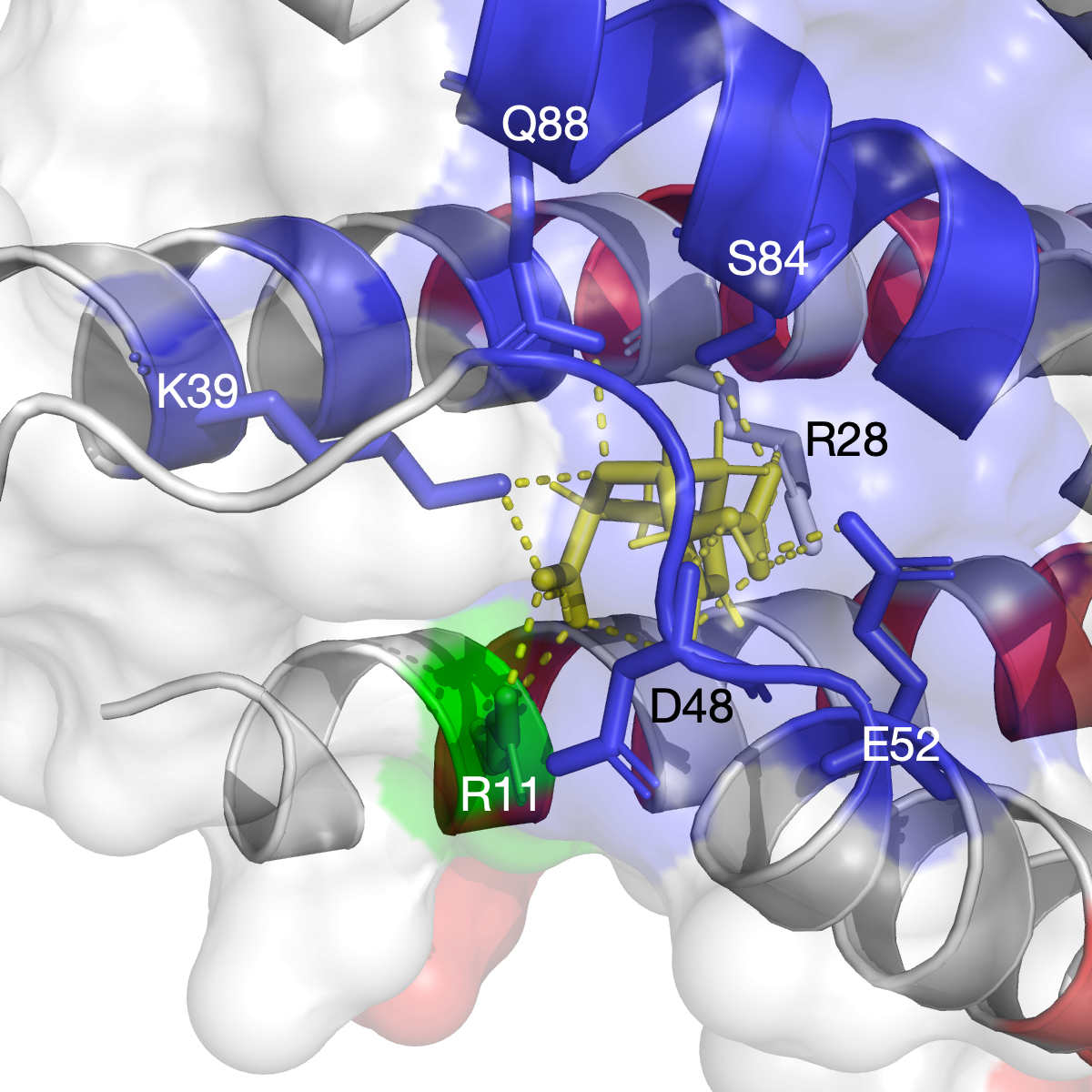
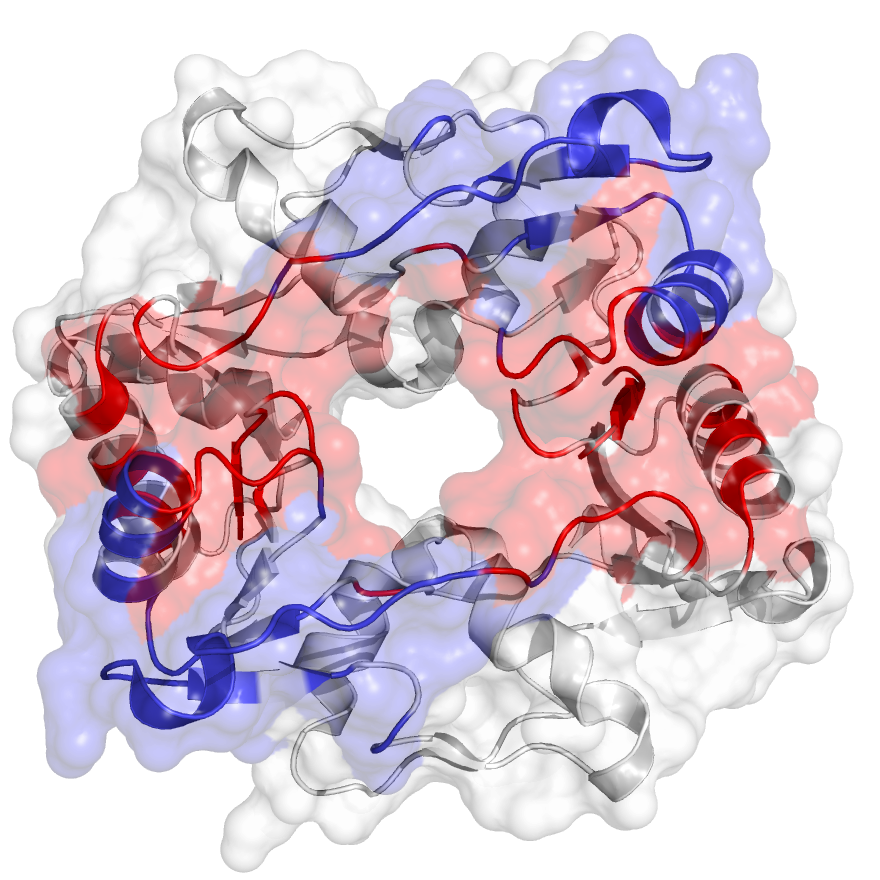
The hinge domain of Smchd1
SMC flexible hinge domain containing 1 (SMCHD1) is a noncanonical member of the SMC protein family, and it has distinct features, e.g., an N-terminal gyrase, Hsp90, histidine kinase, and MutL (GHKL)–type ATPase domain and a hinge domain located at the C-terminus. Disruption of SMCHD1 function is a potential factor that links to human disorders: the muscular dystrophy, facioscapulohumeral dystrophy, and the developmental disorder, Bosma arhinia micropthalmia syndrome. Our method has successfully identified functional communities that are functionally relevant to the canonical dimer interfaces of SMCHD1 hinge domain (PDB: 1GXK).
2020-06-30
Residue communities, functional networks